Saccharomyces cerevisiae Surface Display Vector Kit
More Discount, please inquiry!
Product Description
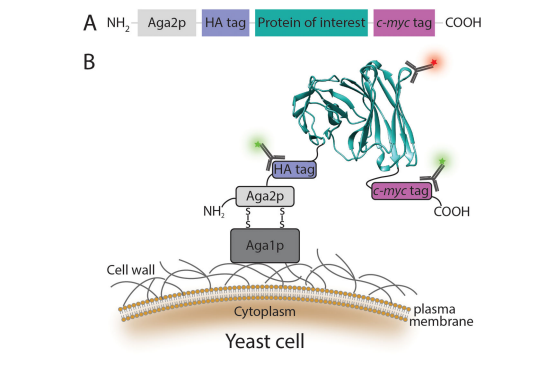
The advanced Saccharomyces cerevisiae Surface Display can be used for applications such as studying protein interactions, antibody screening, and biological material display. The most common yeast display system employs a fusion of the protein of interest to the C-terminus of the a-agglutinin mating protein Aga2p subunit, a technology pioneered by Boder and Wittrup. The yeast surface display construct designed for this system includes two epitope tags: a hemagglutinin (HA) tag between Aga2p and the N-terminus of the protein of interest, and a C-terminal c-myc tag. Induction of protein expression results in surface display of the fusion protein through disulfide bond formation of Aga2p to the β1,6-glucan-anchored Aga1p domain of a-agglutinin. The epitope tags allow quantification of fusion protein expression, and thus normalization of protein function to expression level by flow cytometry using fluorescently labeled antibodies. However, the detection of epitope tags yields no information on the fold or function of the protein of interest. Therefore, a ligand or antibody specific to the native fold of the displayed protein must be used to interrogate these properties.
To increase throughput, yeast-displayed libraries of greater than 108 variants can first be screened using bead-based magnetic-activated cell sorting (MACS) to reduce the library diversity before screening with FACS. In this method, magnetic beads are coated with a soluble target of interest, for example, an antibody or ligand. The beads are then incubated with the yeast library, and yeast displaying non-binding protein variants are removed by washing, after which the yeast binding the desired target is eluted and recovered. This method is advantageous for removing truncated, misfolded, and weak affinity proteins from the library, thereby reducing library diversity to a size that is amenable to quantitative screening by FACS.